Minian
With increasing interest in miniscopes as a method for in vivo calcium imaging, there is need for tools that increase accessibility to the computationally complex process of analysing calcium signals from raw video data. There are several barriers keeping new users from using miniscopes in their research, some of which include understanding the effects of parameters within these analyses, the specialized hardware required to run memory intensive programs, and programming expertise. Zhe Dong and colleagues have thus created Minian to mitigate these challenges.
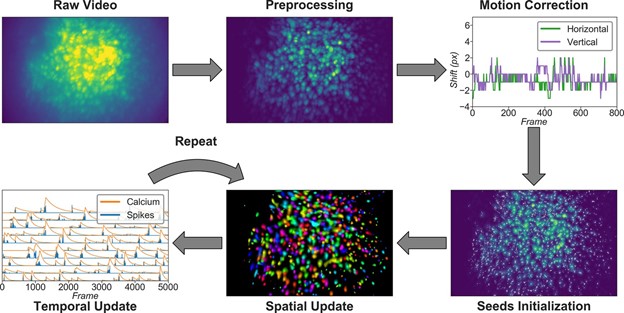
Minian is a user friendly miniscope analysis pipeline that requires low memory and computational demand such that it can be run without specialized hardware. Minion offers interactive visualization that allows users to see how parameters in each step of the pipeline affect the output. This functionality allows users with little computational knowledge to produce high quality calcium imaging results. They also provide detailed documentation and have validated their pipeline across multiple brain regions and neuron types. Minian is thus a great entry point for labs looking to implement miniscopes that are unsure how to choose the best parameters for their specific use case.
This research tool was created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the tool was described, and include an RRID in the Materials and Methods of your future publications. Minian SCR_022601.
Read the Minian Paper
Check out the eLife paper here!
Minian GitHub site
Get access to the code on the GitHub page!