HERBS: Histological E-data Registration in rodent Brain Spaces
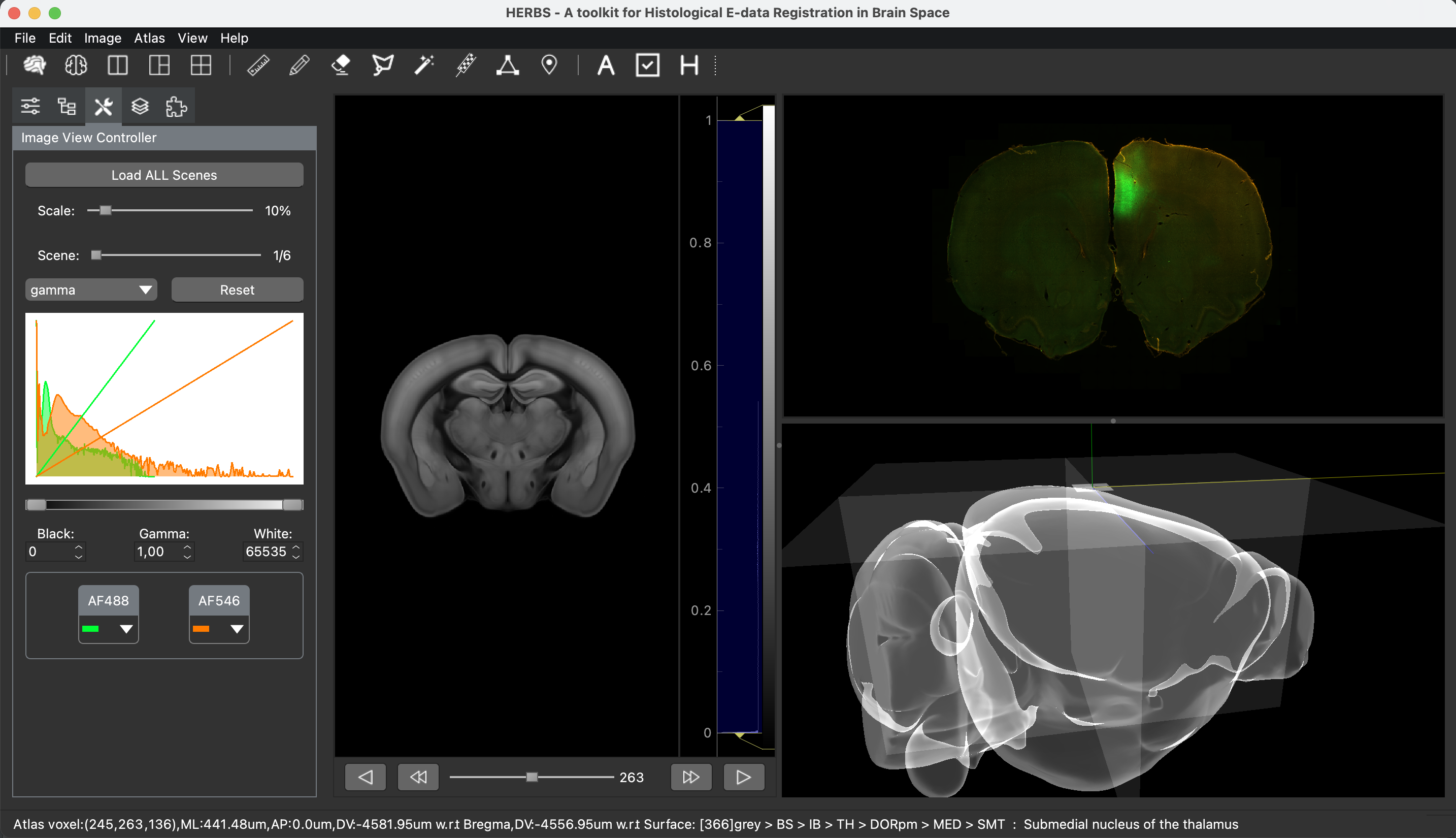
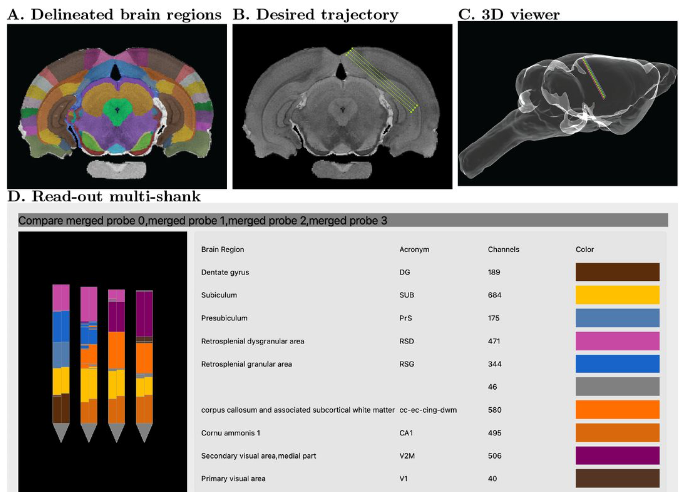
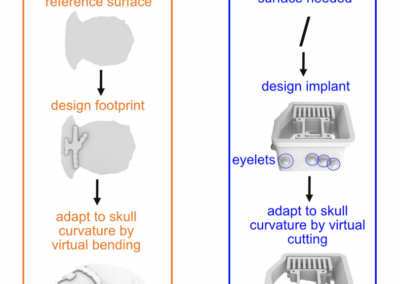
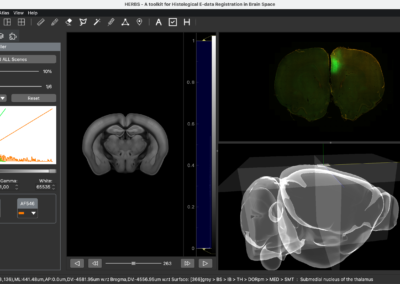
One of the keys to understanding neural correlates of behavior is to understand neural coordinates that we study with various probes. Traditional paper atlases have served as a key tool for researchers both mapping plans for experiments and assessing the histological placement of neural probes, lesions, or virus/protein expressions for decades. However, these atlases are limited to 2 dimensions when trying to conceptualize locations across a rostral to caudal (when viewing coronal images) or medial to lateral (when viewing saggital images) axis. With the advancement of the probes we use to study the brain, we’ve also seen advancements in digital atlases for these purposes. Many of the earlier digital atlases were either limited in 2D like the paper atlases, or required some degree of coding experience for the advanced atlases. To address these issues, Jingyi Guo Fuglstad and colleagues developed and shared HERBS, comprehensive tool for rodent users that offers a broad range of functionalities through a user-friendly graphical user interface written in python. HERBS is based on the Waxholm Sprague Dawley Rat Brain Atlas or the Allen Mouse Brain Atlas, depending on the species of choice. This tool can be used to plan experiments or register recording electrode locations (e.g. Neuropixels, tetrodes), viral expression or other anatomical features, and visualize the results in 2D or 3D. It is compatible with most tissue staining methods and image formats, as long as the regions of interest are visible and can be delineated by the user. The HERBS python GUI is available on PyPI and Github and is compatible with Windows, macOS and Linux systems.
This research tool was created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the tool was described, and include an RRID in the Materials and Methods of your future publications. RRID:SCR_022776
Read the Preprint
Read more in their recent preprint on bioRxiv!
GitHub
Software and downloads from the source!
Check out projects similar to this!