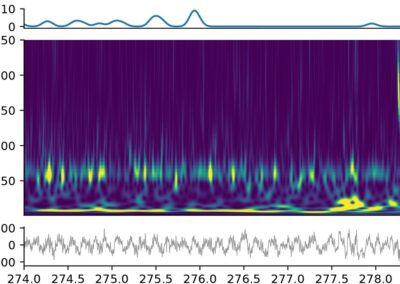
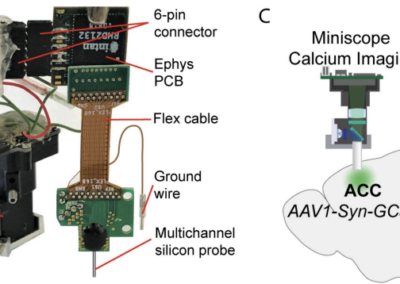
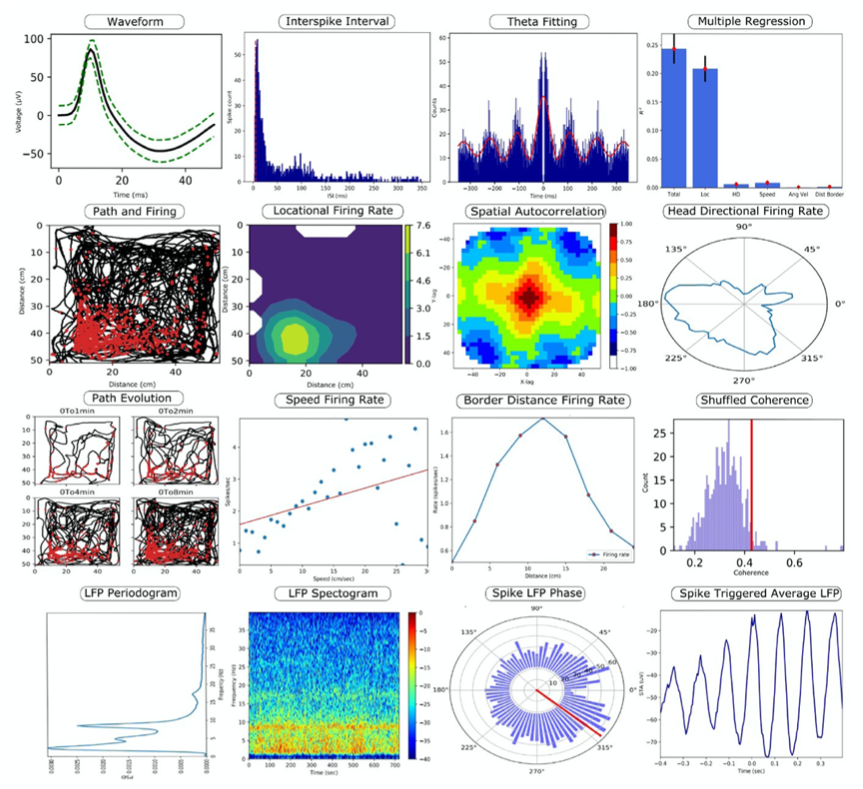
The continuous advancement of technology over the years have helped the neuroscience community begin to understand the complex relationship that exists between the electrical activity in the brain obtained through in vivo recordings (in the form of spikes, spatial, and non-spatial data) and different aspects of an animal’s behavior. Many laboratories today tend to work with custom analysis software due to the scarcity of free-access tools that can aid with recording and processing neuronal information from research with living specimens. Furthermore, most of the software currently available for processing of neurophysiology and behavior data are dedicated to one specific type of data, making it trickier for researchers to record and evaluate different kinds of electrical data from an in-vivo specimen in one run. Under such conditions, scientists require multiple software suites, each one taking care of an individual activity. The use of multiple software suites for different types of data mentioned also results in different laboratories saving their data in different formats, complicating the sharing and editing of data with other laboratories. Md Nurul Islam and his team aim to expand the number of data analysis resources that are freely available for neuroscientists by developing the Neuron Characterization Toolbox (NeuroChaT). The main ambition of the developers of NeuroChaT is to provide the scientific community with the common neurophysiology toolbox by merging into one package a number of different methods/techniques to measure, record, and process multiple kinds of neuronal activity data and their relationship to animal behavior. The developers of this toolbox approach some of the mentioned lab issues by: 1) adoption of the HDF5 format as a default for saving data, as it is a common format that many other programming languages are capable of working with, facilitating the sharing of results with fellow scientists. 2) This toolbox operates via a GUI which makes it pretty easy to use for scientists with little to no experience with programming. Scientists who are comfortable with coding can go ahead and play with its application programming interface (API), this feature will be useful for scientists trying to work with personalized parameters of data. Read more about NeuroChaT and get access to necessary files on GitHub. Read more about NeuroChaT from the original publication from Wellcome Open Research. This post was brought to you by Kevin Chavez Lopez. This project summary is a part of the collection from neuroscience undergraduate students in the Computational Methods course at American University.NeuroChaT
Read the Documentation
NeuroChaT Publication
Thanks Kevin!
Have questions? Send us an email!