SpikeForest
Hot off the eLife press, Jeremy Magland and colleagues have shared SpikeForest, a tool for validating automated neural spike sorters.
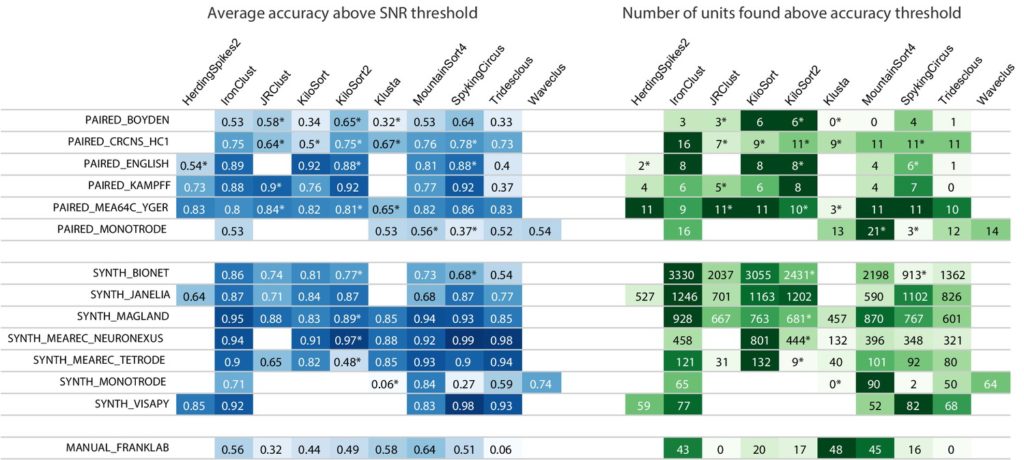
Spike sorting is a crucial step in neural data analysis. Manual spike sorting is time consuming and sensitive to human error, so much effort has been placed into developing automated algorithms to perform this necessary step. However, even with rapid development and sharing of these tools, there is little information to guide researchers for which algorithm may best serve their needs and that it offers the accuracy needed to give a complete scope of the data. To address this, Magland and colleagues across 11 research groups have developed and contributed data for SpikeForest. This python based software suite utilizes a large database of ephys recordings featuring ground truth units (units that have spike patterns known a priori), a parallel processing pipeline to benchmark algorithm performance, and a web interface for users to explore results. This tool can be used to assess which algorithm works best to extract data from different recording and experimental methods (in vivo, ex vivo, tetrode, etc) and provides accurate evaluation metrics for comparison. Information about the spike sorting algorithms that SpikeForest can compare are available in the recent publication, as well as a preliminary comparison of these algorithms based on community provided datasets. The SpikeForest Interface also allows users to sort their own data with a few modifications to the code, which is discussed in the publication. Be sure to check it out!
This research tool was created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the tool was described, and include an RRID in the Materials and Methods of your future publications. Project portal RRID:SCR_021472; Software RRID:SCR_021532
SpikeForest Website
Check out the incredible documentation for this project and learn more about the algorithm on the SpikeForest website.

Read the paper
Read about the project and the algorithm in eLife.