SpikeInterface

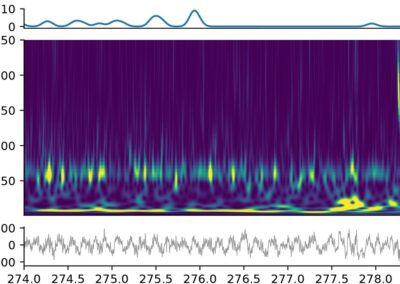
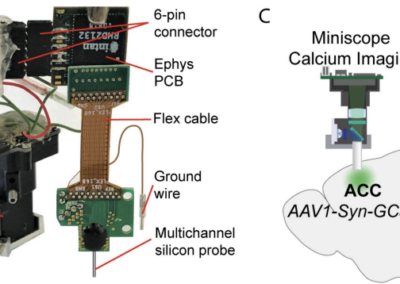
Collecting neural activity is one thing; cleaning, sorting, and analyzing neural data is a whole other beast. In the case of extracellular electrophysiology, there are numerous steps to preprocess data to obtain single unit, or spike activity. With the rapid adoption of high-density neural probes have come an abundance of pipelines, algorithms and software for spike sorting which can make selecting the optimal sorting software somewhat cumbersome and can lead to issues with reproducibility and benchmarking. To address this, Alessio P Buccino, Cole L Hurwitz, and colleagues developed SpikeInterface, a Python framework designed to unify preexisting spike sorting technologies into a single codebase. The goal of this software is to increase the accessibility and standardization of modern spike sorting technologies; make spike sorting pipelines fully reproducible; make data access and analysis both memory and computation-efficient; encourage the sharing of datasets, results, and analysis pipelines; and supply the most comprehensive suite of benchmarking capabilities available for spike sorting. With a few lines of code, researchers can reproducibly run, compare, and benchmark most modern spike sorting algorithms; pre-process, post-process, and visualize extracellular datasets; validate, curate, and export sorting outputs; and more. The Python based API and GUI allow users without coding experience to access a variety of preexisting tools (including Klusta, Mountainsort4, Kilosort, Kilosort2, SpyKING Circus, HerdingSpikes2, Tridesclous, IronClust, Wave clus, and HDsort; as of publication in Nov 2020). SpikeInterface is also compatible with a number of data formats (Klusta, Mountainsort, Phy*, Exdir, MEArec, Neurodata Without Borders, NIX, SHYBRID, Neuroscope, Biocam HDF5, and Binary; as of publication in Nov 2020). The corresponding eLife publication provides a detailed overview of the framework, how it can be utilized, and gives examples of benchmarking and sorting on a widely available Allen Brain dataset. SpikeInterface can be installed through PyPi and full documentation can be found on the SpikeInterface Wiki or Github. The SpikeInterface WIki also includes well-documented tutorials for users to become familiar with the outstanding features of this framework. Be sure to look at it!
This research tool was created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the tool was described, and include an RRID in the Materials and Methods of your future publications. RRID:SCR_021150
Read the Publication!
Read more in their recent eLife article.
SpikeInterface Wiki
Get access to instructions and tutorials for SpikeInterface from the Wiki!