
Jul 11, 2025 | In the Community, OpenBehavior Team Update, Resources, Uncategorized, Video Analysis
This week’s post was written by Ray Shen, an undergraduate student from the University of Maryland who is working with the OB team this summer. Real-Time ArUco Marker Detection in Bonsai-RX ArUco markers are fiducial markers widely used for camera pose...

May 16, 2025 | In the Community, OpenBehavior Team Update, Uncategorized, Video Analysis
This summer, the OpenBehavior project team is focusing on creating documentation for setting up and using three valuable video analysis tools in neuroscience research. These tools often lack straightforward, in-lab setup guides, so we want to make them more...

May 2, 2024 | Community Call to Action, In the Community, Open Source Resource, Resources, Uncategorized, Video Analysis
We recently launched a repository of 3D printed tools for neuroscience research (in August 2022) and started a new GitHub repository to make the designs available in a single repository last month. Thirty five designs are now available in the repository, which you can...
Jul 7, 2022 | In the Community, Open Source Resource, OpenBehavior Team Update, Teaching, Video Analysis
Last year, OpenBehavior created a video repository with the intention of gathering raw behavioral neuroscience videos in one location to train students to use video analysis software. Recently, we have updated our repository and added three videos depicting commonly...
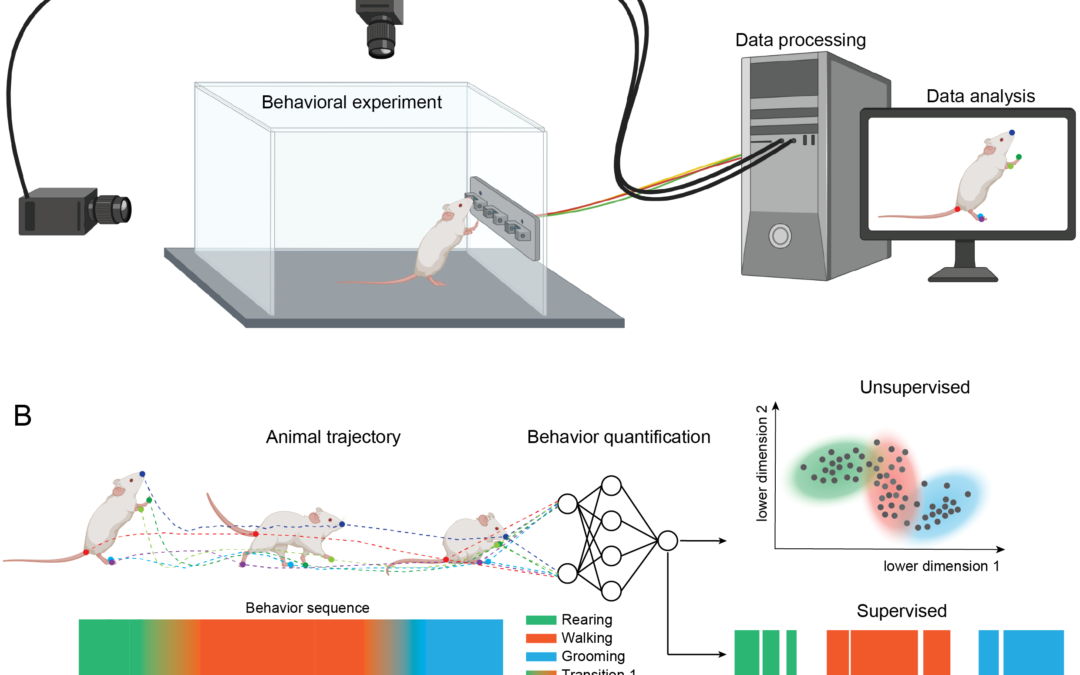
Apr 7, 2022 | Community Call to Action, Open Source Resource, OpenBehavior Team Update, Video Analysis
In the summer of 2021, OpenBehavior organized a working group on methods for video analysis. A Slack channel was started to discuss the methods and a series of virtual meetings were held that involved conversations among users and developers. Some in the group decided...
Sep 2, 2021 | In the Community, Open Source Resource, OpenBehavior Team Update, Teaching, Video Analysis
Earlier this year, the OpenBehavior project initiated a repository of raw videos from typical behavioral neuroscience experiments. A total of 14 video collections were contributed to the repository, and several more will be added this fall. The videos will be useful...





