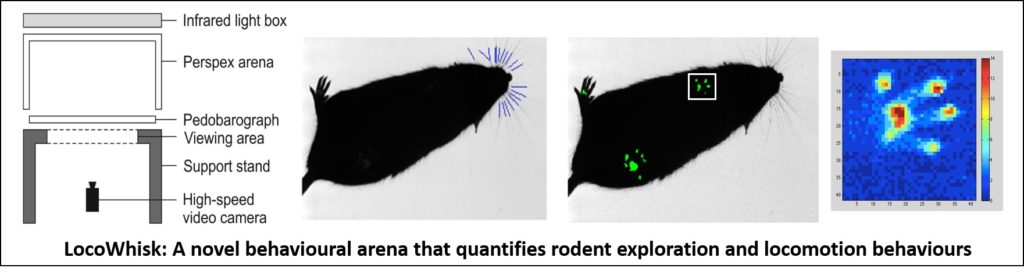
NOVEMBER 21, 2019
Tim Bussey and Lisa Saksida from Western University and the BrainsCAN group developed touchscreen device chambers that can be used to measure rodent behavior. While the touchscreens themselves are not an open-source device, we appreciate the open-science push for creating a user community, performing workshops and tutorials, and data sharing. Most notably, their sister project, MouseBytes, is an open-access database for all cognitive data collected from the touchscreen-related tasks:
Touchscreen History:
In efforts to develop a cognitive testing method for rodents that would optimally reflect a touchscreen testing method in humans, Bussey et al., (1994, 1997a,b) developed a touchscreen apparatus for rats, which was subsequently adapted for mice as well. In short, the touchscreens allow for computer-aided graphics to be presented to a rodent and the rodent can make choices in a task based on which stimuli appear. The group published a “tutorial” paper detailing the behavior and proper training methods to get rats to perform optimally using these devices (Bussey et al., 2008). Additionally, in 2013, three separate Nature Protocols articles were published by this group, with details on how to use the touchscreens in tasks assessing executive function, learning and memory, and working memory and pattern separation in rodents (Horner et al., 2013; Mar et al., 2013; Oomen et al., 2013).
Most recently, the group has developed https://touchscreencognition.org/ which is a place for user forums, discussion, training information, etc. The group is actively doing live training sessions as well for anyone interested in using touchscreens in their tasks. Their twitter account, @TouchScreenCog, highlights recent trainings as well. Through developing automated tests for specific behaviors, this data can be extrapolated across labs and tasks.

MouseBytes:
Additionally, MouseBytes is an open-access database where scientists can upload their data to, or can analyze other data already collected from another group. Not only does this reduce redundancy of experiments, but also allows for transparency and reproducibility for the community. The site also performs data comparison and interactive data visualization for any data uploaded onto the site. There are also guidelines and video tutorials on the site as well.
Nature Protocols Tutorials:
Horner, A. E., Heath, C. J., Hvoslef-Eide, M., Kent, B. A., Kim, C. H., Nilsson, S. R., … & Bussey, T. J. (2013). The touchscreen operant platform for testing learning and memory in rats and mice. Nature protocols, 8(10), 1961.
Mar, A. C., Horner, A. E., Nilsson, S. R., Alsiö, J., Kent, B. A., Kim, C. H., … & Bussey, T. J. (2013). The touchscreen operant platform for assessing executive function in rats and mice. Nature protocols, 8(10), 1985.
Oomen, C. A., Hvoslef-Eide, M., Heath, C. J., Mar, A. C., Horner, A. E., Bussey, T. J., & Saksida, L. M. (2013). The touchscreen operant platform for testing working memory and pattern separation in rats and mice. Nature protocols, 8(10), 2006.
Original Touchscreen Articles:
Bussey, T. J., Muir, J. L., & Robbins, T. W. (1994). A novel automated touchscreen procedure for assessing learning in the rat using computer graphic stimuli. Neuroscience Research Communications, 15(2), 103-110.
Bussey, T. J., Padain, T. L., Skillings, E. A., Winters, B. D., Morton, A. J., & Saksida, L. M. (2008). The touchscreen cognitive testing method for rodents: how to get the best out of your rat. Learning & memory, 15(7), 516-523.
You can buy the touchscreens here.
Editor’s Note: We understand that Nature Protocols is not an open-access journal and that the touchscreens must be purchased from a commercial company and are not technically open-source. However, we appreciate the group’s ongoing effort to streamline data across labs, to put on training workshops, and to provide an open-access data repository for this type of data.